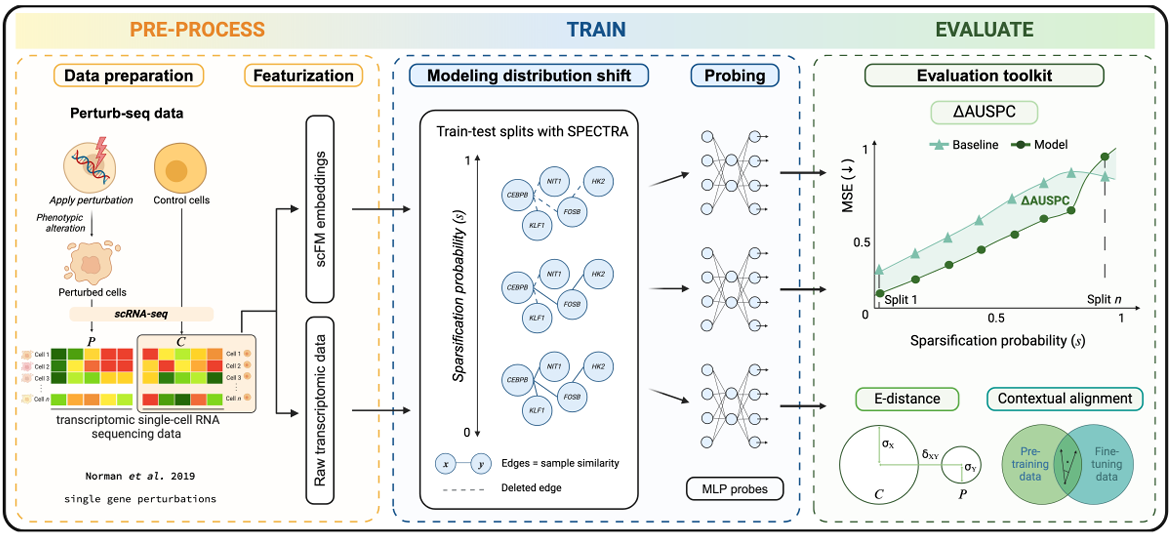
PertEval-scFM: Benchmarking Single-Cell Foundation Models for Perturbation Effect Prediction
2024 | biorxiv | Wenteler A. et al. | Article
In silico modeling of transcriptional responses to perturbations is crucial for advancing our understanding of cellular processes and disease mechanisms. We present PertEval-scFM, a standardized framework designed to evaluate models for perturbation effect prediction. We apply PertEval-scFM to benchmark zero-shot single-cell foundation model (scFM) embeddings against simpler baseline models to assess whether these contextualized representations enhance perturbation effect prediction.
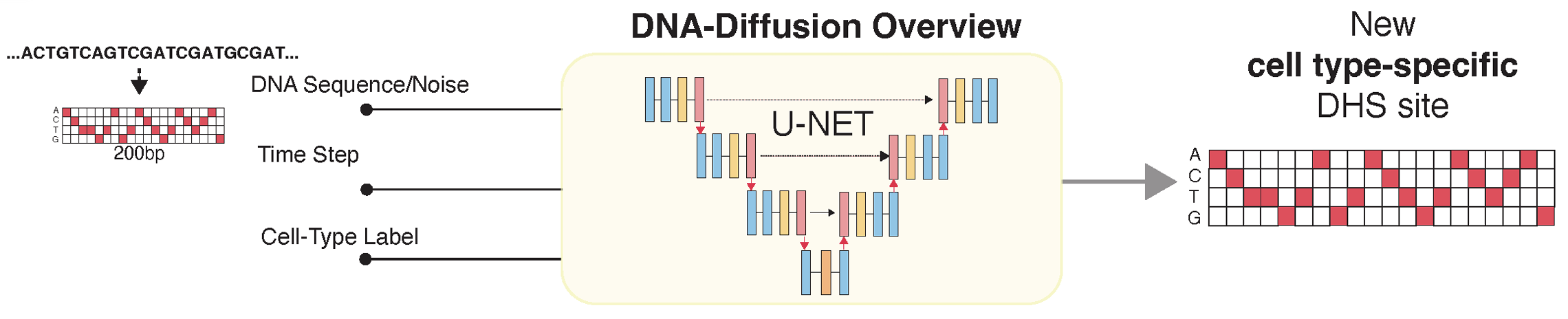
DNA-Diffusion: Leveraging Generative Models for Controlling Chromatin Accessibility and Gene Expression via Synthetic Regulatory Elements
2023 | biorxiv | DaSilva L.F. & Senan S. et al. | Article
The challenge of systematically modifying and optimizing regulatory elements for precise gene expression control is central to modern genomics and synthetic biology. We leverage diffusion models to design context-specific DNA regulatory sequences, which hold significant potential toward enabling novel therapeutic applications requiring precise modulation of gene expression. Our framework uses a cell type-specific diffusion model to generate synthetic 200 bp regulatory elements based on chromatin accessibility across different cell types.
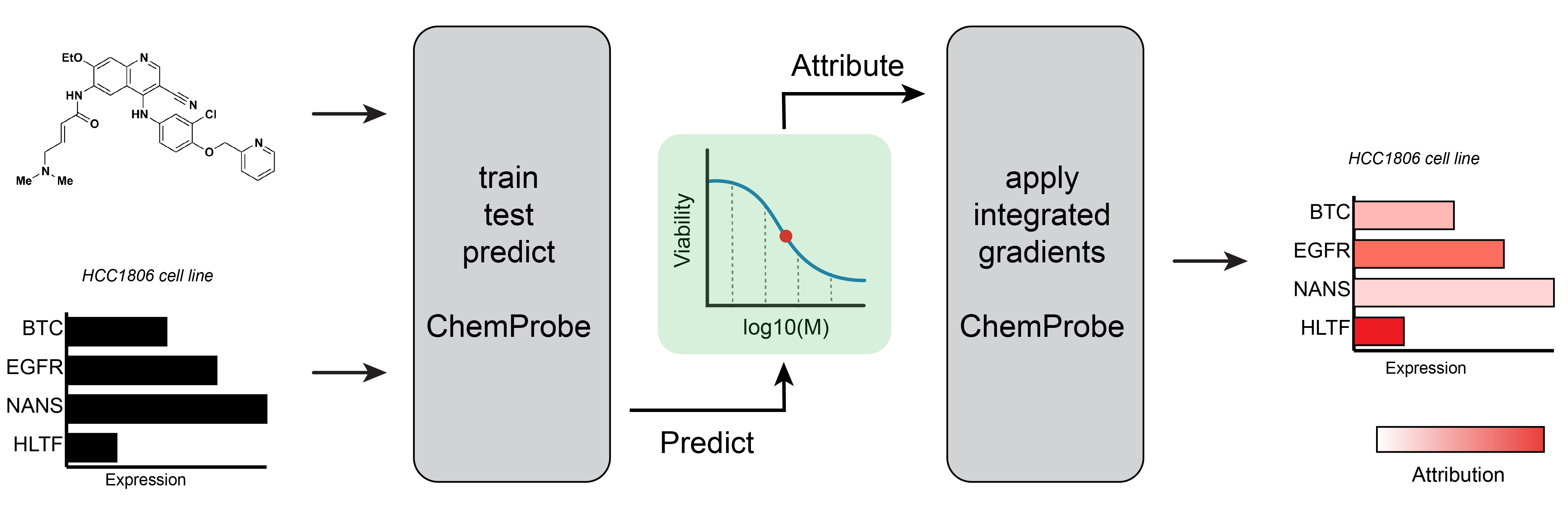
Learning chemical sensitivity reveals mechanisms of cellular response
2024 | Commun. Biol. | Connell W.T., Garcia K., Goodarzi H., Keiser M.J. | Article
Chemical probes interrogate disease mechanisms at the molecular level by linking genetic changes to observable traits. However, comprehensive chemical screens in diverse biological models are impractical. To address this challenge, we developed ChemProbe, a model that predicts cellular sensitivity to hundreds of molecular probes and drugs by learning to combine transcriptomes and chemical structures. Using ChemProbe, we inferred the chemical sensitivity of cancer cell lines and tumor samples and analyzed how the model makes predictions. ChemProbe is an interpretable in silico screening tool that allows new measurements of cellular response to diverse compounds, facilitating research into molecular mechanisms of chemical sensitivity.
A single-cell gene expression language model
2022 | Learning Meaningful Representations of Life, NeurIPS | Connell W.T., Khan U., Keiser M.J. | Article
Given the difficulty of physically mapping mammalian gene circuitry, we require new computational methods to learn gene regulatory rules. Our model, Exceiver, is trained across a diversity of cell types using a self-supervised task formulated for discrete count data, accounting for feature sparsity. Our work provides a framework to model gene regulation on a single-cell level and transfer knowledge to downstream tasks.
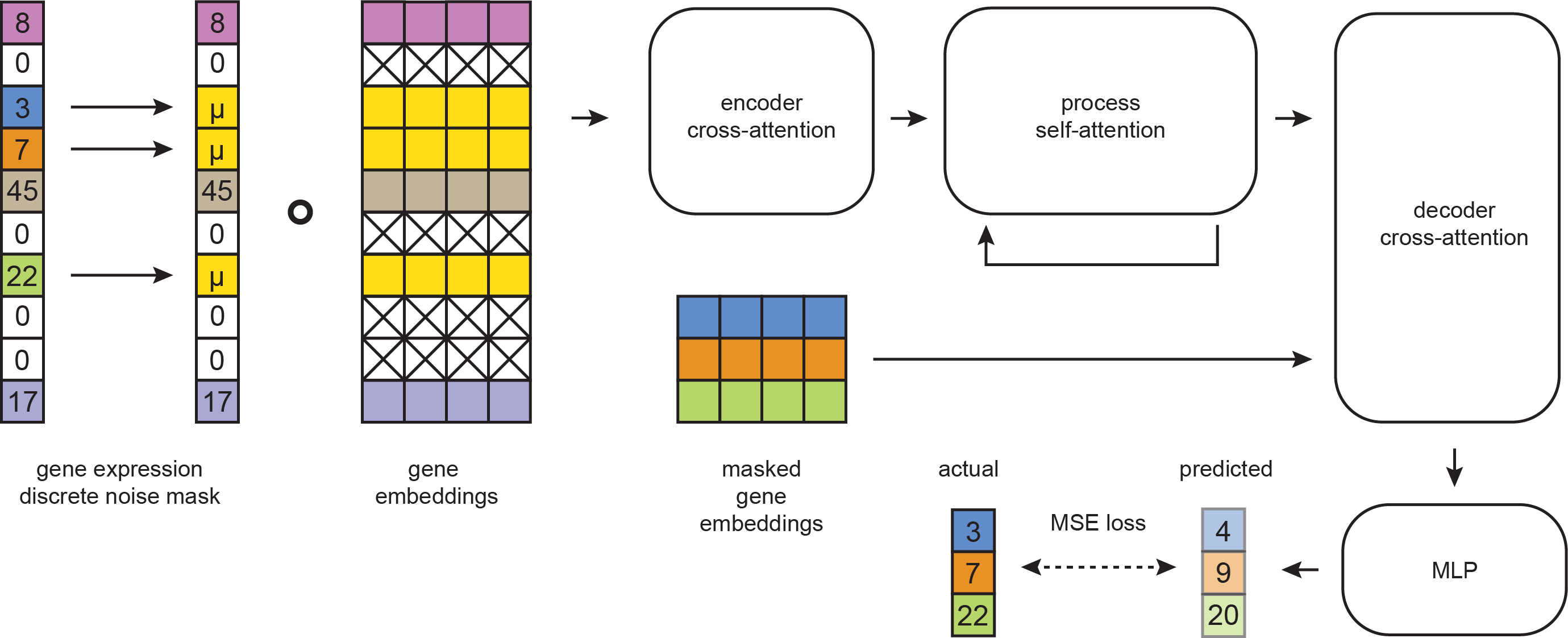
Genome-Wide Association Study of Ustekinumab Response in Psoriasis
2022 | Frontiers in Immunology | Connell W.T., Hong J., Liao W. | Article
Heterogeneous genetic and environmental factors contribute to the psoriasis phenotype, resulting in a wide range of patient response to targeted therapies. Here, we investigate genetic factors associated with response to the IL-12/23 inhibitor ustekinumab in psoriasis. Through GWAS, we identified a novel SNP that is potentially associated with response to ustekinumab in psoriasis.
Nurturing diversity and inclusion in AI in Biomedicine through a virtual summer program for high school students
2022 | PLOS Computational Biology | Oskotsky T. et al | Article
Artificial Intelligence (AI) has the power to improve our lives through a wide variety of applications, many of which fall into the healthcare space; however, a lack of diversity is limiting how broadly AI can impact people. This work can guide AI training programs aspiring to engage and educate students entirely online, and motivate people in AI to strive towards increasing diversity and inclusion in this field.

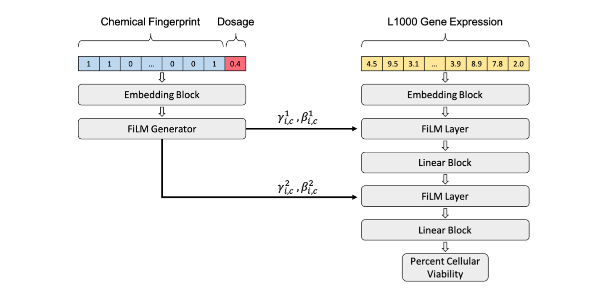
Predicting Cellular Drug Sensitivity using Conditional Modulation of Gene Expression
2020 | Learning Meaningful Representations of Life, NeurIPS | Connell W.T., Keiser M.J. | Article
Selecting drugs most effective against a tumor’s specific transcriptional signature is an important challenge in precision medicine. To assess oncogenic therapy options, cancer cell lines are dosed with drugs that can differentially impact cellular viability. This ongoing work formalizes in silico cellular screening as a conditional task in precision oncology applications that can improve drug selection for cancer treatment.
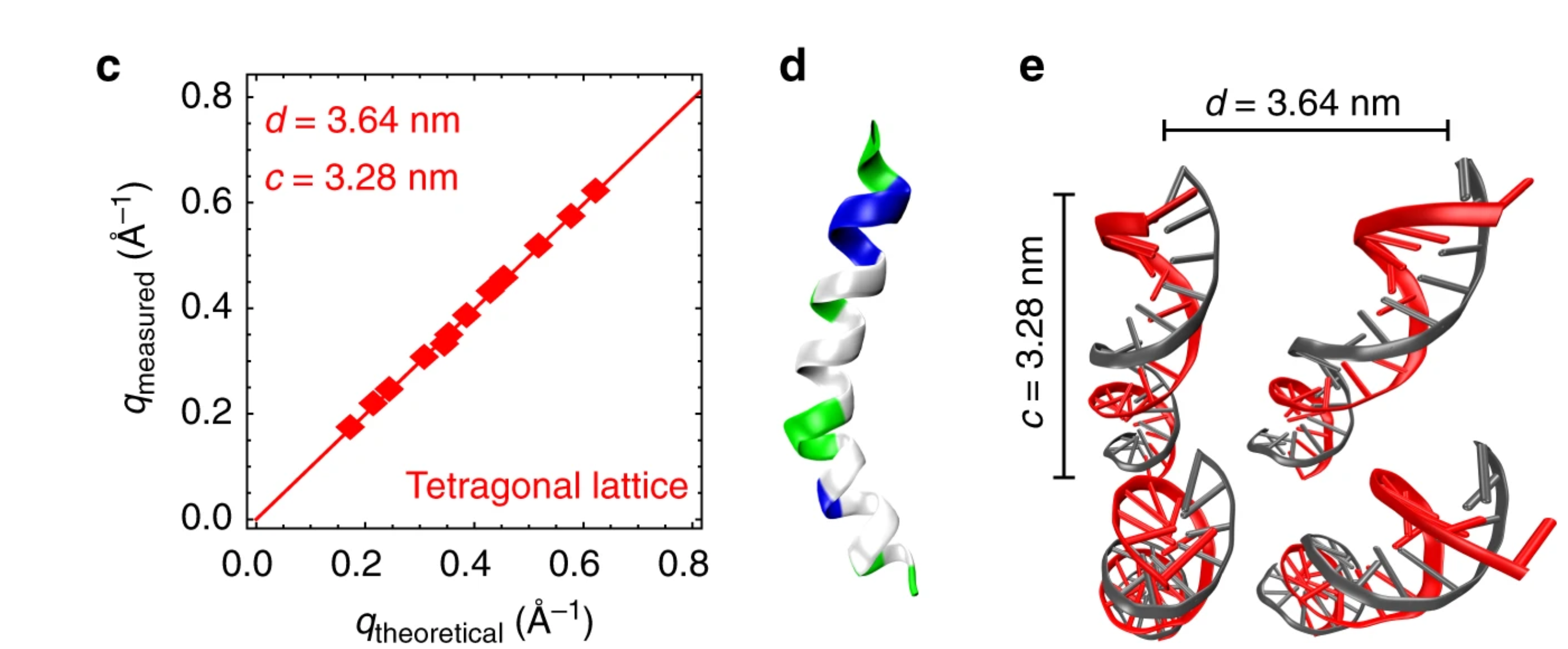
Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9
2019 | Nature Communications | Lee E., Zhang C., Di Domizio J., Jin F., Connell W., Hung M., Malkoff N., Veksler V., Gilliet M., Ren P., Wong G.C.L. | Article
Amphiphilicity in ɑ-helical antimicrobial peptides (AMPs) is recognized as a signature of potential membrane activity. Some AMPs are also strongly immunomodulatory: LL37-DNA complexes potently amplify Toll-like receptor 9 (TLR9) activation in immune cells and exacerbate autoimmune diseases. The rules governing this proinflammatory activity of AMPs are unknown. Here we examine the supramolecular structures formed between DNA and three prototypical AMPs using small angle X-ray scattering and molecular modeling.
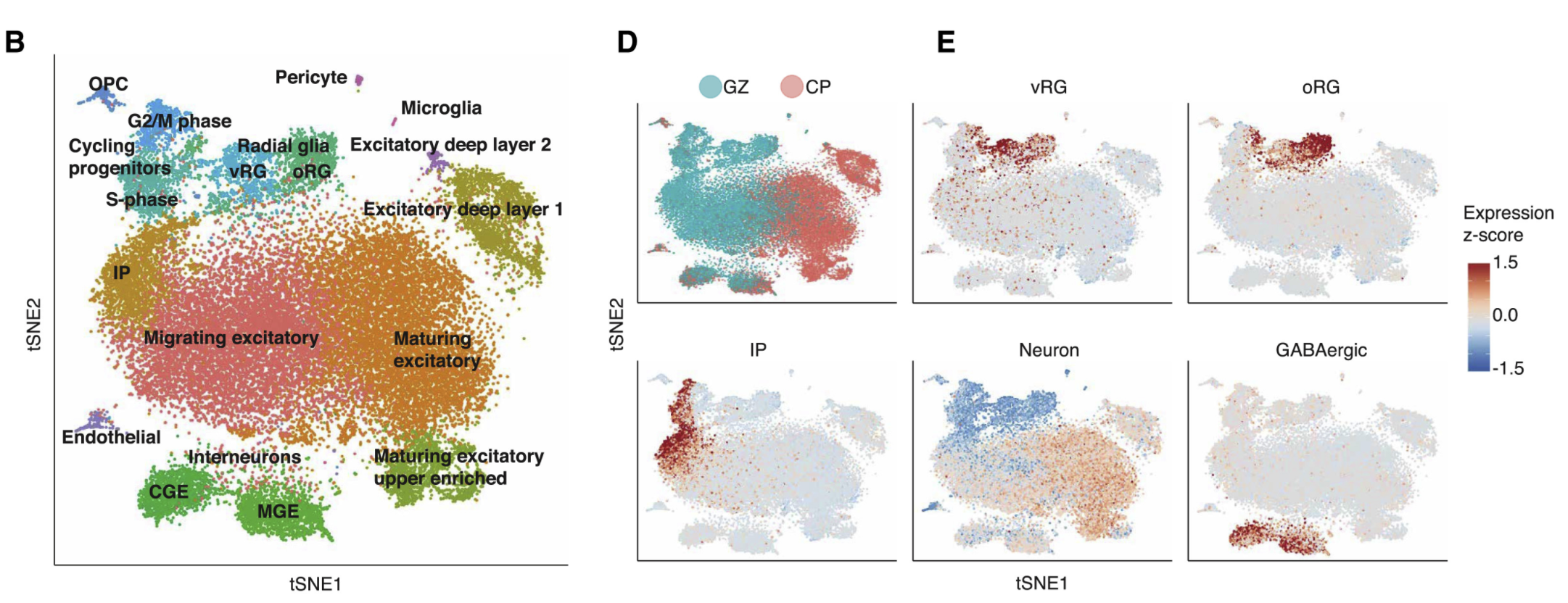
A Single-Cell Transcriptomic Atlas of Human Neocortical Development during Mid-gestation
2019 | Neuron | Polioudakis D., Torre-Ubieta L., Langerman J., Elkins A.G., Shi X., Stein J.L., Vuong C.K., Nichterwitz S., Gevorgian M., Opland C.K., Lu D., Connell W., Ruzzo E.K., Lowe J.K., Hadzic T., Hinz F.I., Sabri S., Lowry W.E., Gerstein M.B., Plath K., Geschwind D.H. | Article
We performed RNA sequencing on 40,000 cells to create a high-resolution single-cell gene expression atlas of developing human cortex, providing the first single-cell characterization of previously uncharacterized cell types, including human subplate neurons, comparisons with bulk tissue, and systematic analyses of technical factors. These data permit deconvolution of regulatory networks connecting regulatory elements and transcriptional drivers to single-cell gene expression programs, significantly extending our understanding of human neurogenesis, cortical evolution, and the cellular basis of neuropsychiatric disease.